My name is Eldad Klaiman, and I'm a Software Engineering Manager and Engagement Lead at Roche Information Solutions. Today, I'm going to be talking about how Roche is leveraging open source mentality and developer communities through its journey of digitalization, in order to deal with new types of algorithm developments in computer vision.
About Roche Information Solutions
Roche Information Solutions is a relatively new thing. It was founded a couple of years ago, with the aim to be a software engineering-oriented organization within Roche to drive digital technology in a rapidly digitalizing healthcare market.
Research Decision Support is a team inside Roche Information Solutions that develops internal research tools for our internal scientists and developers.
The platforms and tools I'll be sharing with you today aren’t commercially available, but they're being used by a growing community of scientists and researchers inside the company.
We're working with oncology data sets, which are largely computer vision data sets for digital pathology and radiology images. Although our platforms are focused on the oncology disease area, they can also be extended to other disease areas that have imaging modalities.
And lastly, the engagement team, which I lead, is responsible for the advanced users, features, and support. Advanced users are basically what we call our internal algorithm developers who are coding and developing new algorithms.
The three stages of pharma digitalization
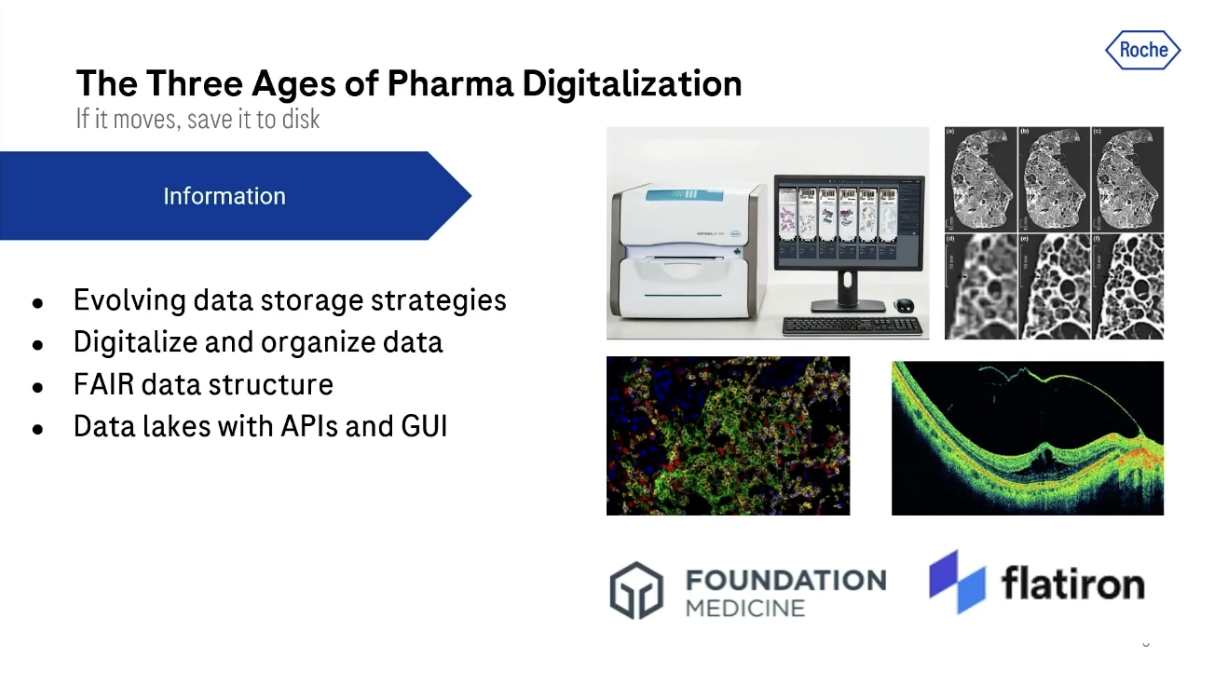
Stage 1: Information
I was lucky enough to join Roche at the beginning of its digitalization journey about 10 years ago. Initially, we digitalized everything. We got the slides from microscopes, scanned them, and stored them on disks. We got different patient metadata, and those were then stored.
We were basically working with tens and hundreds of Excel sheets with file names and where those were stored and where they came from.
And slowly, as developers and data scientists started using these data sets across the organization, collecting them and using them to develop new applications and algorithms, we saw a shift and an evolution in data storage strategy.
From being files on disks and in folders, these data sets became cloud-based data lakes with APIs and graphic user interfaces that allowed you to search, access, and manage the data.
Concepts like FAIR were also evolving; findability, accessibility, interoperability, and reusability of data. And really, it was pretty amazing to see this kind of evolution of data strategies.
Stage 2: Computation
The next step came with deep learning. This was the evolution and growth of GPU capabilities and a big buzz around deep learning, especially in computer vision with the success of ImageNet.
Roche identified the potential in this technology and started building compute capabilities inside the company. We had several high-performance computing (HPC) clusters built inside the company, and those were available for our data scientists, algorithm developers, and researchers to use freely.
This led to very broad experimentation in this field. We conducted myriads of experiments in fields like semantic segmentation, survival prediction, weakly supervised learning, style transfers, and so on.
This allowed us to also compete in different public challenges and achieve quite a high ranking in those as well.
Stage 3: Consolidation
So we organized the data, we got our data lakes, we got the compute, and we experimented a bit with the different use cases in deep learning. But what now?
I think we've been entering a sort of consolidation stage in this area in the last few years. We're trying to leverage the research that we've done in this field in deep learning and computer vision to try and leverage it to bring more business value to our customers, open up opportunities, and develop different functionalities and products.
We're doing this in personalized healthcare by developing predictive algorithms for patient risk or mortality. We're doing this in companion diagnostics by developing algorithms that detect antigens for specific drugs.
We're also doing this in reverse translation or drug discovery by developing Explainable AI that basically allows us to understand the tumor microenvironment better, and to inform back to our discovery team so that they can create better drugs and molecules.
In order to support this algorithm development internally, we're developing a platform that activates multimodal data. Users can use this platform to develop their algorithms and quickly pass them to the clinical infrastructures.
We're helping data scientists and algorithm developers to develop the algorithms and integrate them.
But before I show you the actual platform itself, let's talk a bit about these algorithm developers that we're supporting.
A new type of algorithm developer has emerged in computer vision
As I said, we work with these algorithm developers to help them develop the algorithms. Traditionally, these algorithm developers have been computer scientists and engineers that were very proficient in coding and computer vision. They knew what the Fourier transform and feature engineering was, and this requires some specific type of support to help these guys develop.
Slowly, with the emergence of deep learning, we see more and more engineers and data scientists developing algorithms for computer vision. However, they have no computer vision domain knowledge because all the software developer needs to do is train a convolutional neural network with some annotated data, and they get the computer vision algorithm without needing to do explicit feature engineering.
And even more concerning than this is in the past year, we have had a new type of algorithm developer, and this is related to the buzz around AI.
I just want to take a quick detour to explain what I think might be one of the root causes that influenced this change in the type of algorithm developer.
Polanyi’s paradox is named after the British-Hungarian philosopher, Michael Polanyi. He basically talks about how human knowledge of our own capabilities and actions is to a large extent beyond our explicit understanding.
This is summarized in the sentence, ‘We know more than we can tell.’ This means that the mundane, daily tasks that we do are mainly based on our intuitive tacit knowledge, and we can't really verbalize them or explain how we do them.
This is very common in many of the things that we do day to day, like driving a car, identifying somebody's face, or lifting a glass of water. It's very hard to explain in detail how to do that.
And this is why for many years, this has been recognized as one of the obstacles in the field of AI and automation; in order to write an algorithm or a system that does something automatically, you need a very good description of how you perform the task.
All of this has now basically changed, but this is why we mostly still see iRobot-type robots and not C-3PO housemaids.
How deep learning has transformed the computer vision developer profile
I've mentioned this change, but deep learning completely sidesteps this whole paradox by helping us train algorithms without needing to explain to them what they should do.
All we really need to do with deep learning is to give it annotated data. So basically data and labels, and recently not even labels.
This has even progressed with AutoML methods by allowing us to develop algorithms without coding at all, without even selecting a network architecture, without specifying hyperparameters, or anything else. All we need is a tonne of data.
This has changed dramatically in computer vision use cases for two main reasons. One is the exponential growth of the amount of data that we have. The imaging data comes from natural sources like cell phones and cameras, and also in the medical field, from high-resolution cities to digital pathology scanners.
So, we have trillions of datasets and pixels that give us a lot of opportunities in this field of deep learning.
Another thing is that these imaging data sets are typically annotated by very expensive experts. These annotations take a lot of time, and it's also very inconsistent.
For example, recent studies show that pathologist concordance in annotations is less than 70%. I've actually conducted experiments at Roche where pathologist concordance was less than 40% in cell classification. This is a big problem.
So essentially, we have our new algorithm developers now.
These clinicians, biologists, and pathologists that have been sitting on tonnes of annotated data that they themselves created, have seen people like me, data scientists, and engineers, use the data that they prepare in order to train these AI and machine learning algorithms, and they suddenly realize that they can do the same thing.
Add to that the fact that in traditional pharma companies, there have always been more biological natural scientists and clinicians than data scientists. This has always caused long waiting times to get your data analyzed or your hypotheses tested.
So basically, we're now facing an influx of new algorithm developers that don't know how to code, don't know anything about computer vision, and barely know anything about deep learning. But they want to develop computer vision algorithms with deep learning. So this makes the work of supporting these algorithm developers much more challenging.
There is one silver lining about all of this. Clinicians are typically very conservative, but because of this buzz around AI, they're willing to be more welcoming of new algorithms now, especially if they're involved in the development of those. And this allows us to introduce new technologies that we weren't able to do before since they were considered experimental or science fiction.
How Roche’s products support the different types of algorithm developers
Our approach leverages structural data lakes with a graphic user interface that allows us to enable non-coding developers to organize their data in different data sets to filter out things.
Then we have a self-serve paradigm with internal open-source libraries that users can leverage in order to develop their specific algorithms. And by delivering the correct tool to each user, we’re minimizing the need for active support.
The IRISe framework
The IRISe framework is basically our data lake. But it's not only a data lake, it's a web-based platform that allows users to design experiments, perform data curation activities, and develop and deploy their algorithms online.
This platform was initially limited to digital pathology data, but because of the popularity of its features and functionality, we’re now expanding it to support clinical imaging like CTs and PET, and other modalities as well.
The platform application is designed as a web portal, and this eliminates the need to install any software on the client's computer. It also removes any dependency on the computer hardware or the operating systems.
Once you're logged in to this platform, you can upload images, organize them into data sets according to your preferences, and copy images from one project to another in order to create the setup and the data sets that you need to train your algorithm.
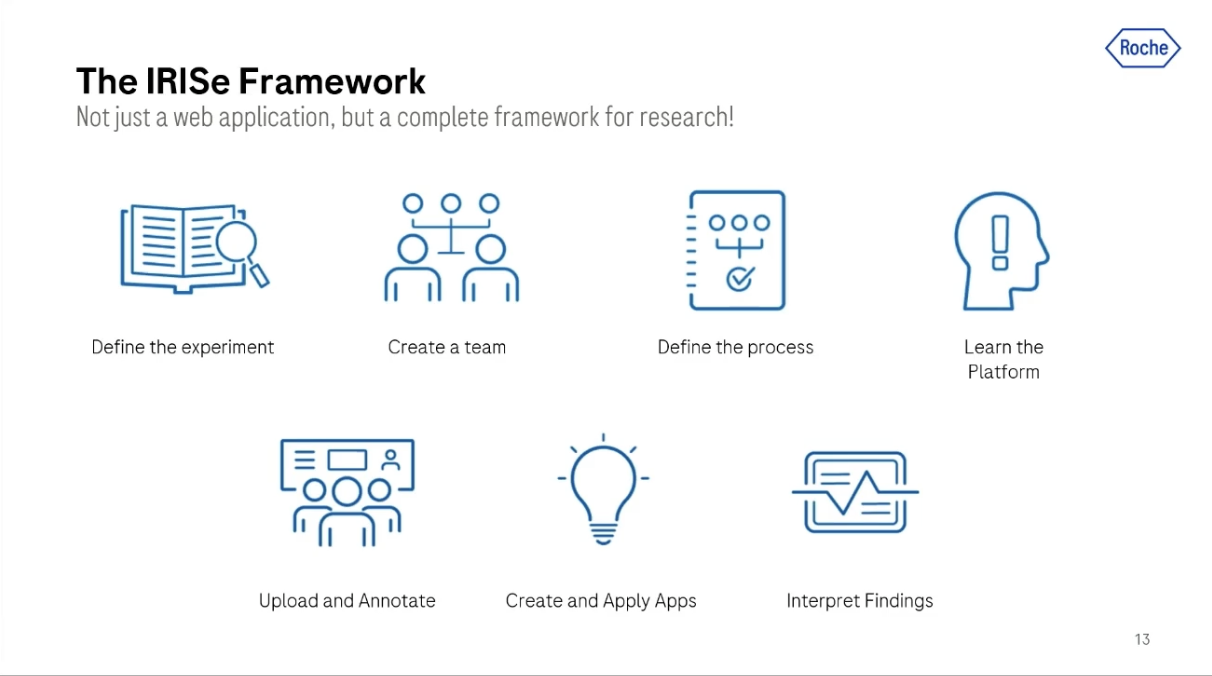
You can also create a team and share those data sets and projects with the different team members.
Once data is organized in this project and experiments, the user can view them in a dedicated web viewer. This viewer supports different types of manual annotations and visualization of algorithm results.
The platform also contains an app catalog to which users can publish their applications in order to share them with the community. They can also trigger applications that they publish there on thousands of slides on our platform compute resources with a click of a button. This makes the algorithms useable for the entire community without needing to think about how or where to run and deploy them.
Open source solutions
‘If you give a man a fish, he will be hungry tomorrow. But if you teach a man to fish, he will be richer forever.’ With this proverb in mind, we’re developing inner source libraries for the advanced user community.
Developers are not only permitted to download and reuse the code, but they're encouraged to fork it, use it wherever they want, copy it, and take it away from us. We encourage them to contribute back to our source code and to our community, and the advantages of doing that are getting support from our team of experts and also from the different community experts.
This kind of community with the open source magnet at the center triggers a lot of collaboration between different departments, silo breaking, and basically creates this community of experts that goes and supports itself.
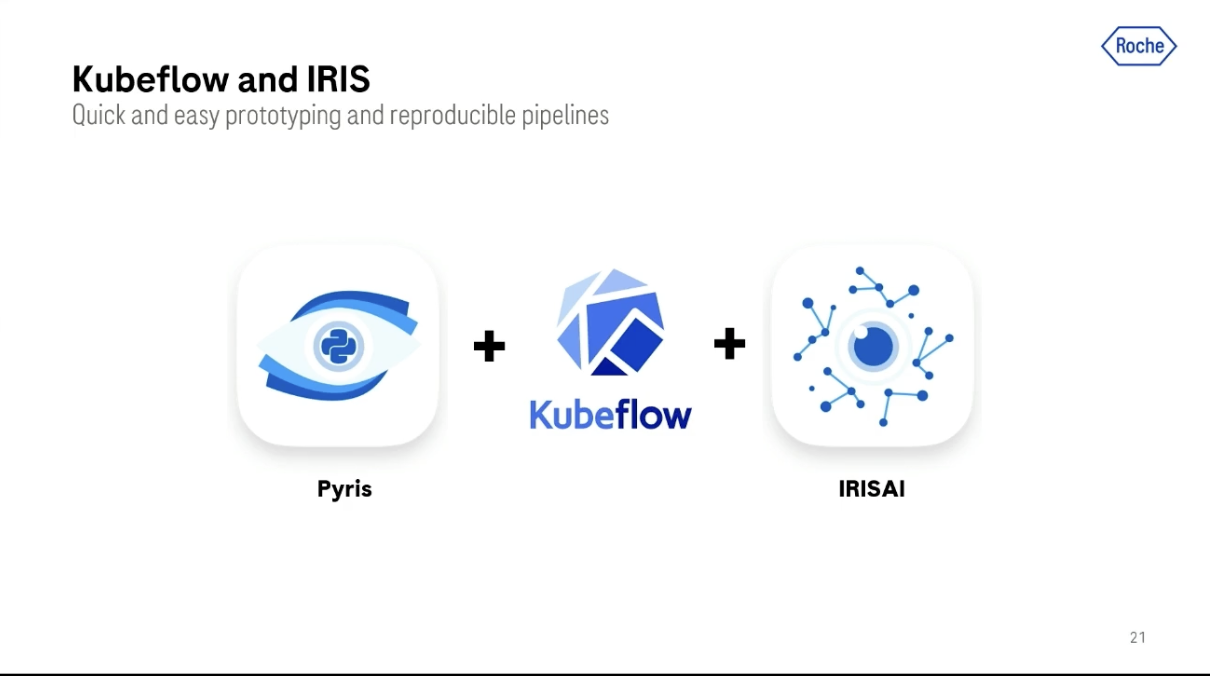
We offer our customers two advanced user open-source products. One of them is Pyris, which is a shallow-level Python API library that allows the user to connect to our platform and interact with the data on a framework.
You can basically use it to push and pull images, post annotations on an image, or upload or download analysis results for a specific image asset.
The other open-source library we offer is called IRISAI, which is basically a portfolio of deep learning-based modules for algorithm development. This was also initially only for pathology, but now we support different use cases and modalities like radiology and genomics. We have modules that are specifically designed for semantic segmentation, survival prediction, multimodal analysis, and others.
When I started developing IRISAI as purely a research tool in pharma, we were two users basically using this software. But in the last two years, as we open-sourced it, the number of advanced users using this product grew from two to 200.
In comparison, the engagement team that supports these user groups grew from three engineers to six.
We then spent the first year after open-sourcing just building tools and methods that’ll allow us to manage this growth and scale up.
We have Slack channels for the community to talk to each other and receive support from our team. We have automatic documentation servers and a website with dedicated blogs that explain the different modules in the library. Each of these blogs contains a runnable Jupyter Notebook that the user can download and run without having any prior knowledge of that specific technology.
Kubeflow
Addressing a growing need in the community for quick and easy prototyping and reproducible pipelines, we're also starting to leverage Kubeflow in order to extend our platform and the services that we provide.
We're doing this first of all with Kubeflow pipelines. Kubeflow pipelines is a module in Kubeflow that allows you to consistently and reproducibly run complex workflows of algorithms.
We have pipeline templates in IRISAI that provide an internal compute environment and allow users to trigger and manage their advanced analytics experiments.
And since Kubeflow pipelines are run with a graphic user interface, non-coding users can also leverage this feature in order to trigger experiments and analysis on datasets.
These pipelines can also be systematically run on schedule in order to run existing algorithms and verify the results on new incoming data sets to see if there’s any data drift.
Another module that we're using in Kubeflow is Kubeflow Notebook Servers. This is a module that originally allowed a user to launch a Jupyter Notebook server on Kubeflow. In the last few years, this feature has evolved to support more and more functionality and run more software like VS Code, and recently customizable user software.
IRIS-Extensions
This enabled us to conceptualize what we now call IRIS-Extensions. IRIS-Extensions are basically interactive web applications that are connected to our platform via our API libraries like Pyris. They can be developed, uploaded, and triggered at will by any user.
One of the things that we noticed is that with the influx of new users with new use cases, we had a lot of new user interface functionality requests. These requests were flooding our engineering teams and creating long development queues.
IRIS-Extensions help us to overcome these queues by allowing users to develop their own user interface by themselves and trigger that connected to our platform data on the Kubeflow framework. This allows us to eliminate these long queues and achieve better result satisfaction as well.
Another advantage of this extension concept is that it allows us to quickly and efficiently connect third-party software to the data on our platforms as well.
DeePathology Studio
One example of such an extension is DeePathology Studio. DeePathology is a company that actually emerged from Roche’s own startup accelerator in Munich. They created software that allows codeless development of digital pathology, with use cases like cell detection, segmentation type, classification, and object detection.
This off-the-shelf web app has been integrated into IRIS through the extension concept, and it allows our customers to create deep learning-based computer vision algorithms and deploy them on our app catalog without writing a single line of code.
All of the images and annotations that we generate using this third-party software are synchronized via Pyris and with our platform data so that we remain with a single point of truth. And this collaboration with the pathology and the concept of various extensions allowed us to complete this integration in record-breaking time with almost no use of our internal engineering resources.
Summary
Deep learning has transformed the types of audiences that want to develop computer vision applications.
Finding ways to service and maintain this large influx of new developers with no software or computer vision domain knowledge is a big challenge for us.
Creating open-source communities and self-service solutions can mitigate the stress on engineering teams and create self-sustaining user ecosystems.



 Follow us on LinkedIn
Follow us on LinkedIn


